
Our Research
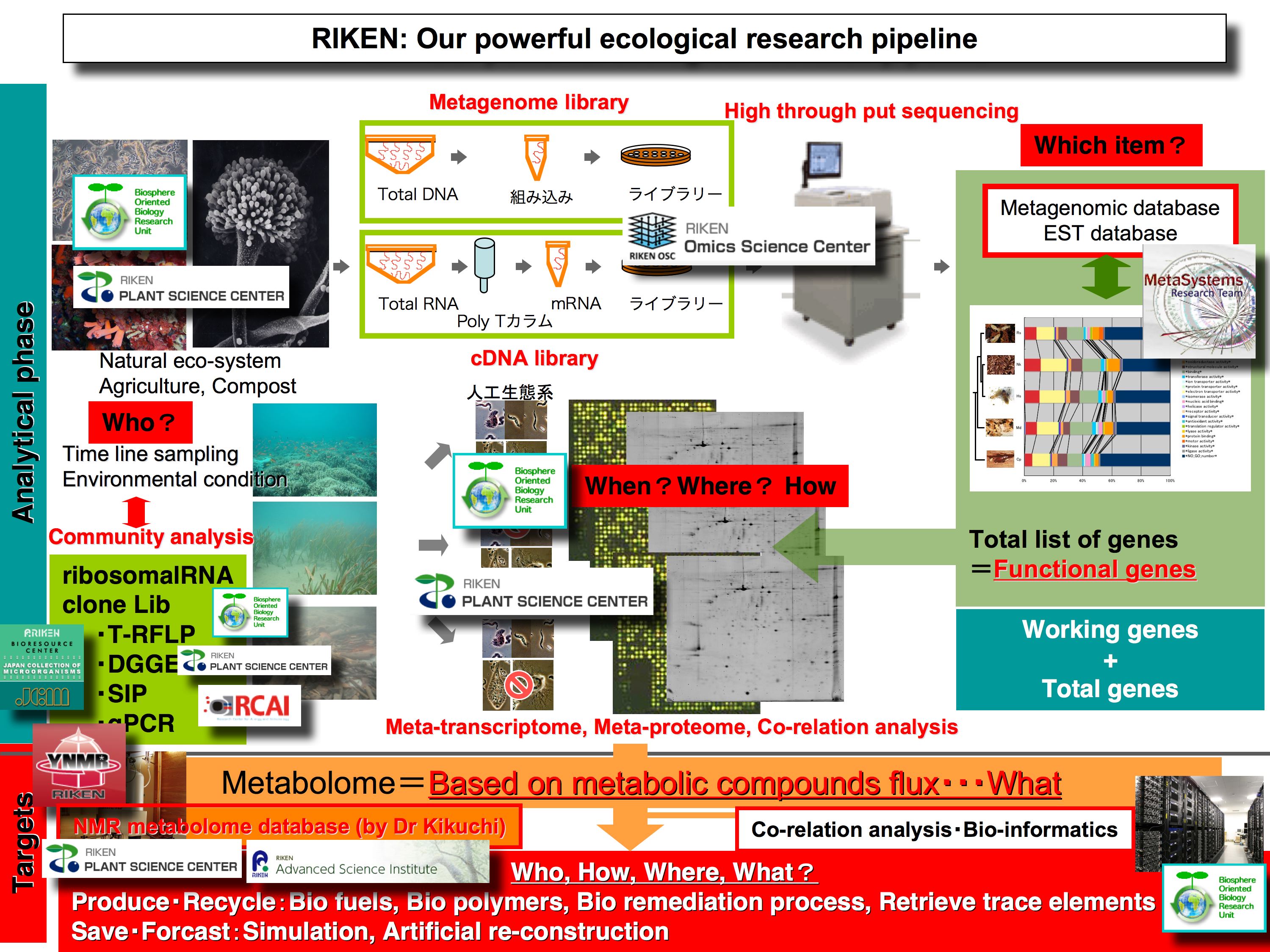
Our unit performs "multi-omics" analyses in order to understand the complexity of ecosystem function and to establish methodologies for investigating the not-yet-cultivated component of microbial communities. Such novel analysis concepts will open a new horizon in biological science and biosphere science.
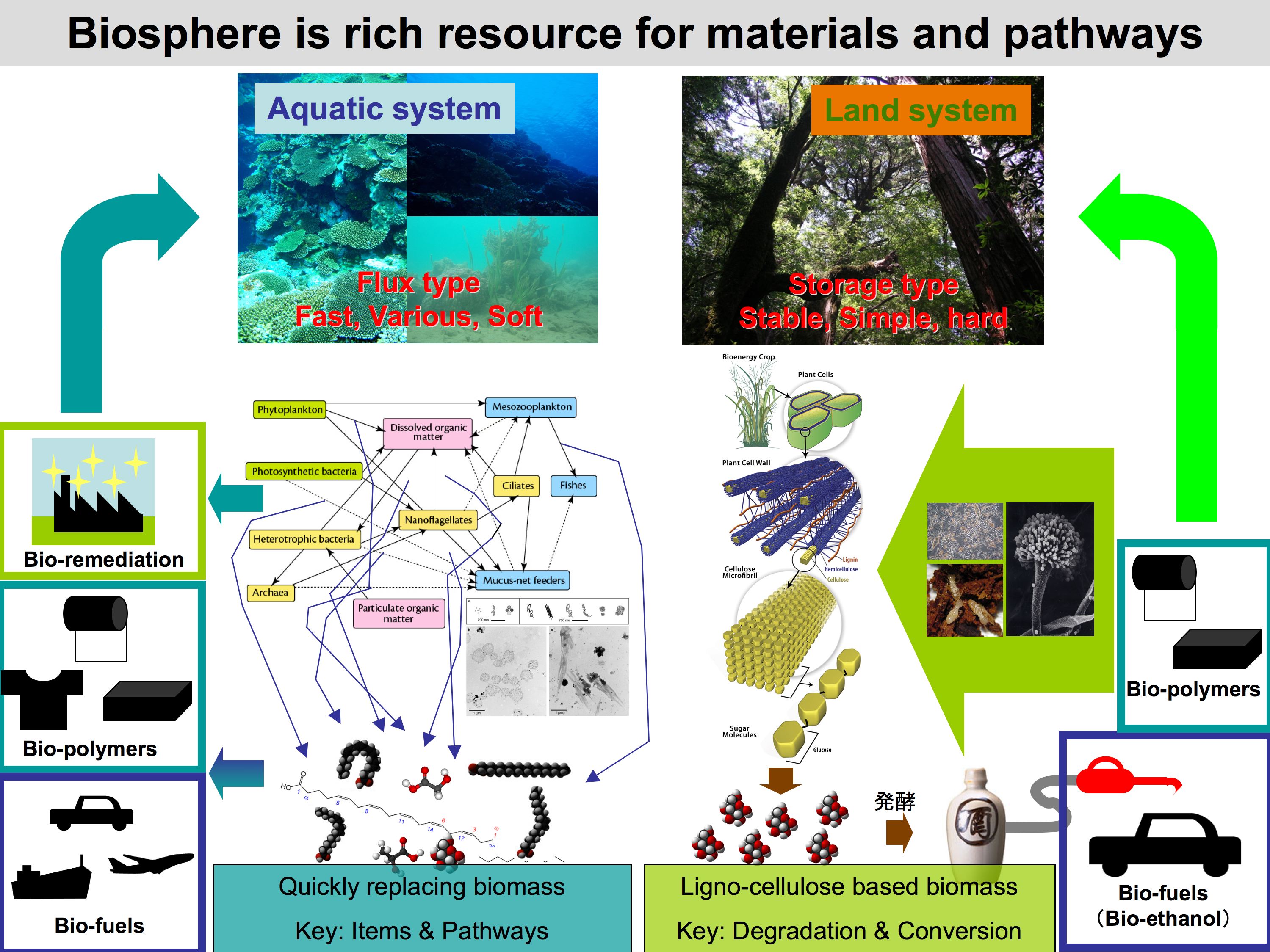
Biomass is one of the most important research targets in biology today. We consume fossil fuel resources and release carbon dioxide leading to global climate change. Recently, first generation bio-fuels produced from crops instead of fossil fuels have shown promise in partially reducing climate change. However, we now face serious conflicts between bio-fuel production and food production.
The biosphere contains various organic materials and energy flow. These components include useful materials for bio-fuel production. However, most organisms that contribute to this huge system are unknown microbes that cannot be cultivated. Globally, and particularly in the United States, many research efforts to develop such bio-resources are underway using a meta-genomic approach.
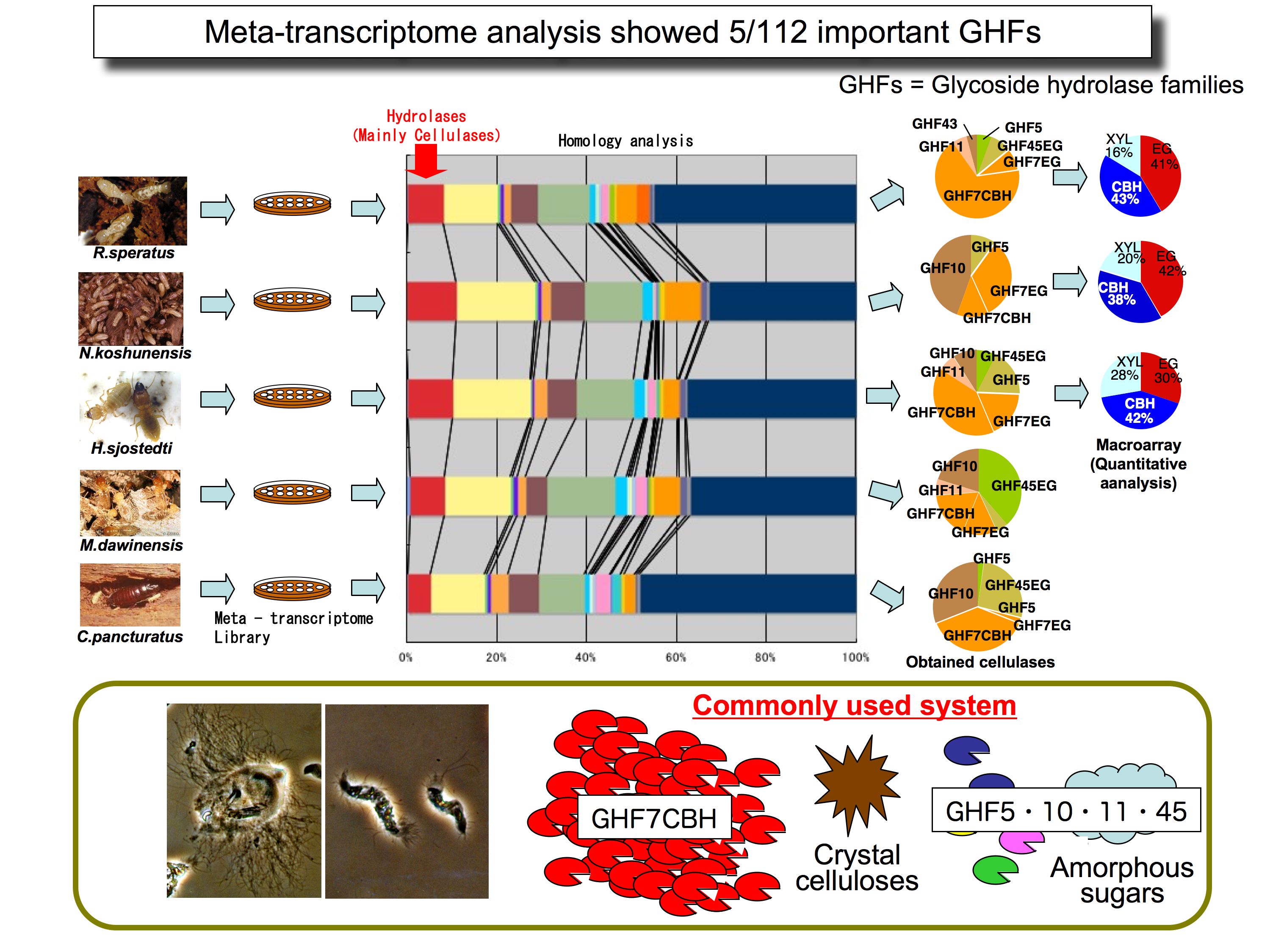
The main target of our unit is the useful microbial resources in a given ecosystem. However, our approach is totally different from a metagenome driven strategy. Our approach is a function / molecule driven strategy. For example we have already obtained novel high-performance cellulases from the termite symbiotic system by using meta-transcriptome methods. These enzymes can realize non-chemical / physical process to produce bio-ethanol and organic compounds for bio-polymer from ligno-cellulose based biomass.
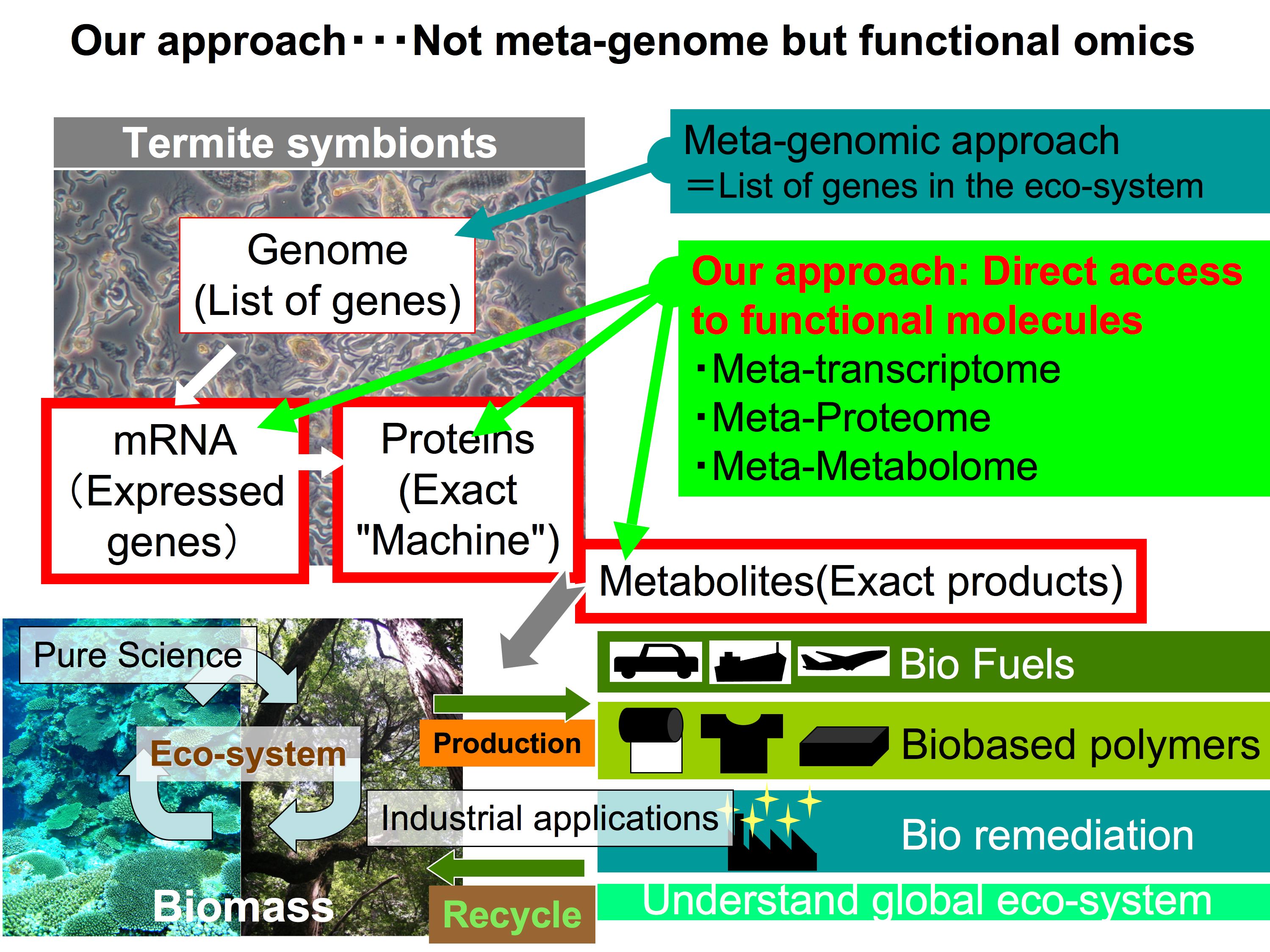
This strategy can be applied to other ecosystems. For example, the marine
phytoplankton community. Currently, we are performing a series of studies to
understanding about this complex / huge ecosystem using our multi-omics strategy.
Our goal is understanding of the function of complex ecosystem for subsequent
utilization to establish basic technology for a sustainable society. We believe that our approach is unique and it will
contribute not only to build the technology for a sustainable society but that it will
also establish a brand new type biology: Biosphere oriented biology.

1.Transcriptome analysis of host termite
Microorganisms dwell symbiotically in the termite hindgut. In this
study we identified genes that contribute to the role of the host in
maintaining the symbiotic relationship with microorganisms. The body
tissue and digestive organs (salivary gland, foregut, midgut, hindgut)
dissected from the lower termite Hodotermopsis sjostedti were used for
the analyses. The transcriptomes in these organs were investigated
using expressed sequence tag (EST) analysis and real-time quantitative
RT-PCR. The cDNA libraries in the salivary gland and foregut included
not only cellulase genes, but also several genes involved in glucose
production, hemi-cellulose degradation, chitin degradation, the innate
immunity system, and anti-microbial activity. Gene expression analyses
by real-time RT-PCR revealed that the genes associated with cellulose
degradation, innate immunity component, and anti-microbial proteins
were much more strongly expressed in the salivary gland than in other
tissues. Our results identify the functional genes used in the host of
the termite symbiotic system.

2.Single cell genomics of symbiotic protist of termite
Single-cell oriented genome analysis of Trichonympha agilis was
performed. Obtained genomic DNA from the single nucleus of Trichonympha
agilis was amplified by the Rolling Circle Amplification (RCA) method and
the DNA was analyzed by high throughput DNA sequencing (FLX).
We obtained 800Mbp FLX data and an additional 80Mbp ordinary sequence data
by capillary sequencer. Results of assembly analysis showed that
obtained contig size was still short. We conclude that the sum of our obtained data
is smaller than the expected genome size of Trichonympha agilis.
We propose two possible working hypotheses, i) Trichonympha
agilis has a polyploid genome or ii) Trichonympha has a huge genome over 100Mbp.
To evaluate these hypotheses, we are now trying to
select fine sequence data, reassemble with this selected data set and perform an "ORF
saturation curve" analysis.

3.Integrated multi-omics analysis of Termite eco-system and their ecological function
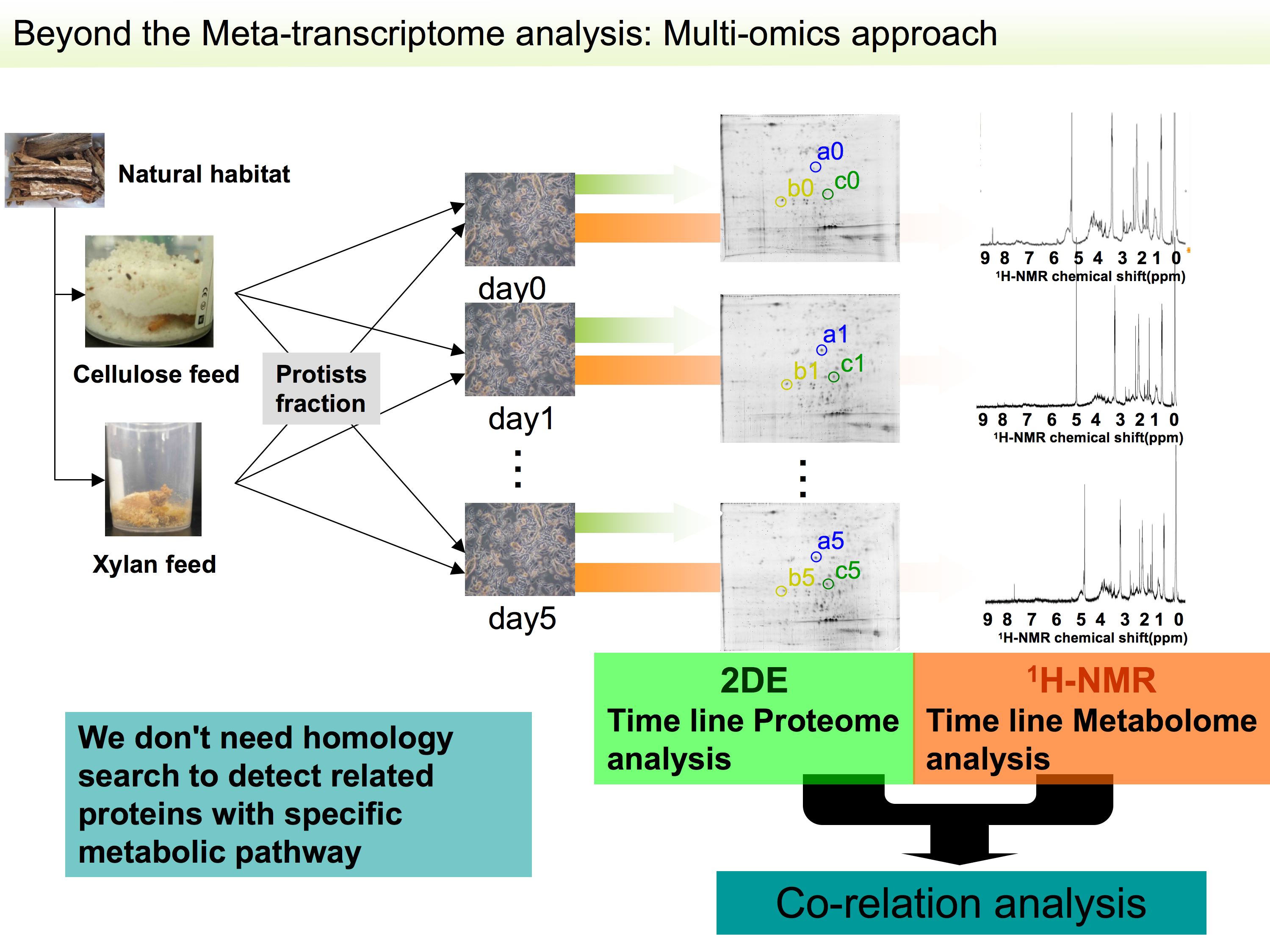
We performed
purification condition setting for the specific and unique cellulolytic
enzyme, Glycosyl Hydrolase Family (GHF) 45 endoglucanase from the symbiotic
protistan community of the termite.
Ion exchange column, Hydrophobic interaction column and Gel filtration
column were evaluated for this purpose. The results indicate that the ion
exchange column can remove main contaminant, amilase, from the starting
material. The hydrophobic interaction column can adsorb the objective enzyme
and the gel filtration column can divide other contaminant proteins.
Therefore, we used these column methods and purified the GHF45 endoglucanase from culture fluid of transgenic Aspergilus
oryzae. Now we are analyzing the character of this unique enzyme from the symbiotic protists of the termite.
We have also set conditions for proteome analysis to obtain novel factor(s) for biomass degradation from symbiotic protists of the
termite.
Results of correlation analysis between timeline metabolome analysis
and proteome analysis with symbiotic protistan fraction of cellulose
fed termites showed that some novel factors related with biomass
degradation and utilization by symbiotic protists of termite include
possible a factor- Cip2-like protein. This peptide can cleave the connection
between hemi-cellulose and lignin. Now we are trying to get
heterogeneous expression of this Cip2-like protein using an E. coli system to
evaluate the contribution of this enzyme to biomass degradation
reactions.

4.Integrated multi-omics analysis of the marine algal ecosystem and ecological function
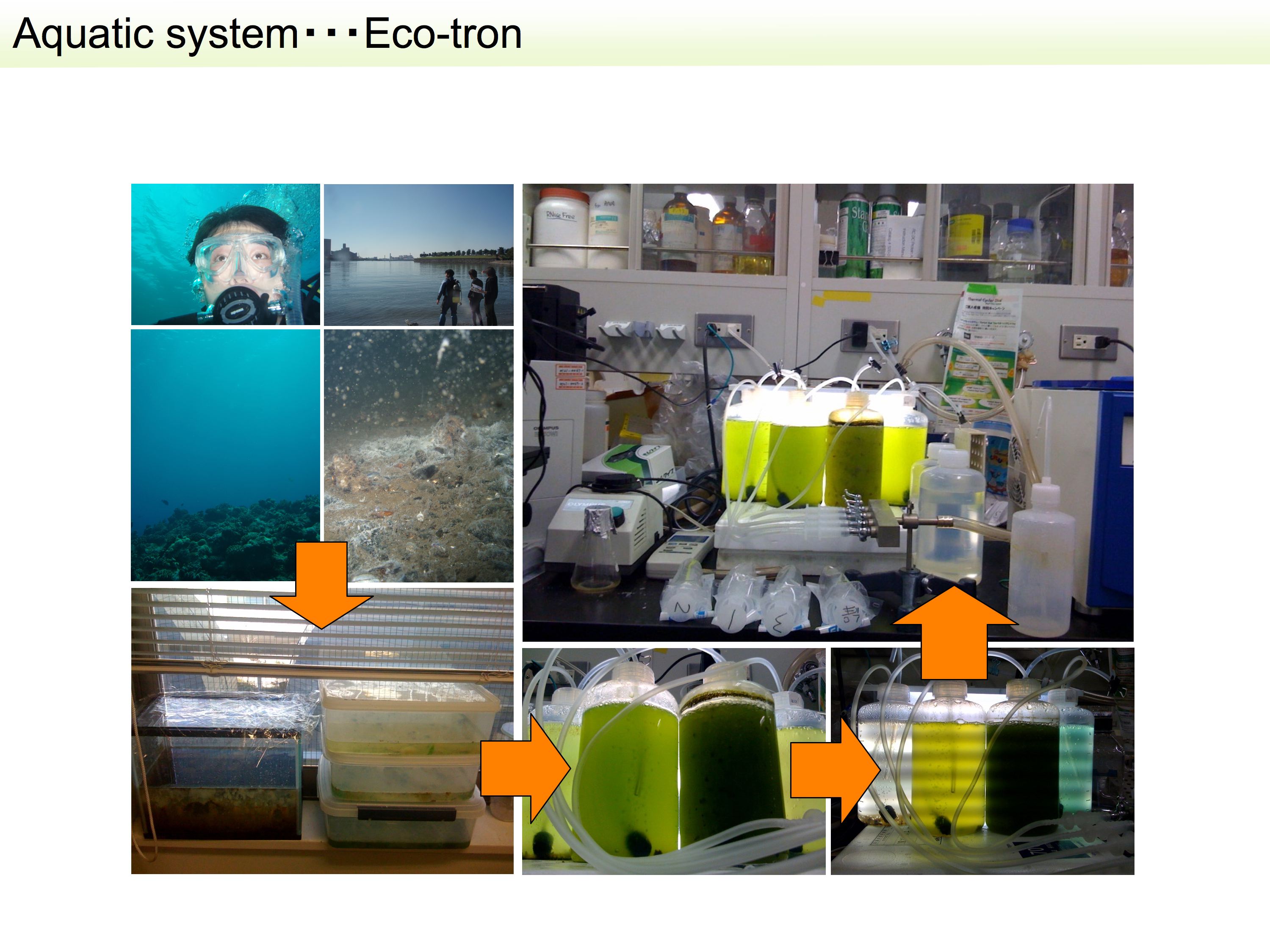 Because of their ease of maintenance in the laboratory, high potential for new discoveries and the potential for direct applications to many research areas, including nutrient cycling, bloom dynamics (including harmful algal blooms), pollution mitigation and carbon-neutral biofuel
production, eutrophic marine ecosystems are an ideal research topic. Although marine phytoplankton play a major role in global carbon cycling, global climate and nitrogen cycling, knowledge about how marine microbial communities function is limited. Major, globally-important groups have only recently been described (e.g. Prochlorococcus in 1992 and eukaryotic biliphytes in 2008), and fundamental questions about the generation and maintenance of phytoplankton blooms and diversity remains largely unresolved. Further, marine plankton show great promise for exploration of new metabolites and compounds; they produce numerous products for defense, signaling, attack, and nutrient storage. Diatoms, green algae and cyanobacteria in particular synthesize abundant lipids and poly-saccharides, which have recently shown promise as biofuel sources. Recent work has also indicated that marine microbial communities are extremely diverse and capable to rapidly respond to environmental
changes, both genetically and physiologically, suggesting great potential for controlled metabolic studies.
Because of their ease of maintenance in the laboratory, high potential for new discoveries and the potential for direct applications to many research areas, including nutrient cycling, bloom dynamics (including harmful algal blooms), pollution mitigation and carbon-neutral biofuel
production, eutrophic marine ecosystems are an ideal research topic. Although marine phytoplankton play a major role in global carbon cycling, global climate and nitrogen cycling, knowledge about how marine microbial communities function is limited. Major, globally-important groups have only recently been described (e.g. Prochlorococcus in 1992 and eukaryotic biliphytes in 2008), and fundamental questions about the generation and maintenance of phytoplankton blooms and diversity remains largely unresolved. Further, marine plankton show great promise for exploration of new metabolites and compounds; they produce numerous products for defense, signaling, attack, and nutrient storage. Diatoms, green algae and cyanobacteria in particular synthesize abundant lipids and poly-saccharides, which have recently shown promise as biofuel sources. Recent work has also indicated that marine microbial communities are extremely diverse and capable to rapidly respond to environmental
changes, both genetically and physiologically, suggesting great potential for controlled metabolic studies.
|








